Overview of protocol
We developed a minimally harmful protocol for extracting DNA from the historical specimens . Our protocol is heavily inspired by the work of Shpak et al., 2023 PLOS Biology. Following an overview of our workflow. For a more detailed protocol see here.

Following some impressions of steps during the protocol with various taxa (including larger ones such as Blattela germanica)

Specimens before and After Treatment
D. immigrans (freshly pinned)
3 examples of specimens before (left) and after (right) DNA extraction with our protocol

D. sucinea (freshly pinned)
3 examples of specimens before (left) and after (right) DNA extraction with our protocol

D. funebris (collected in 1887)
example of a specimen before (left) and after (right) DNA extraction with our protocol

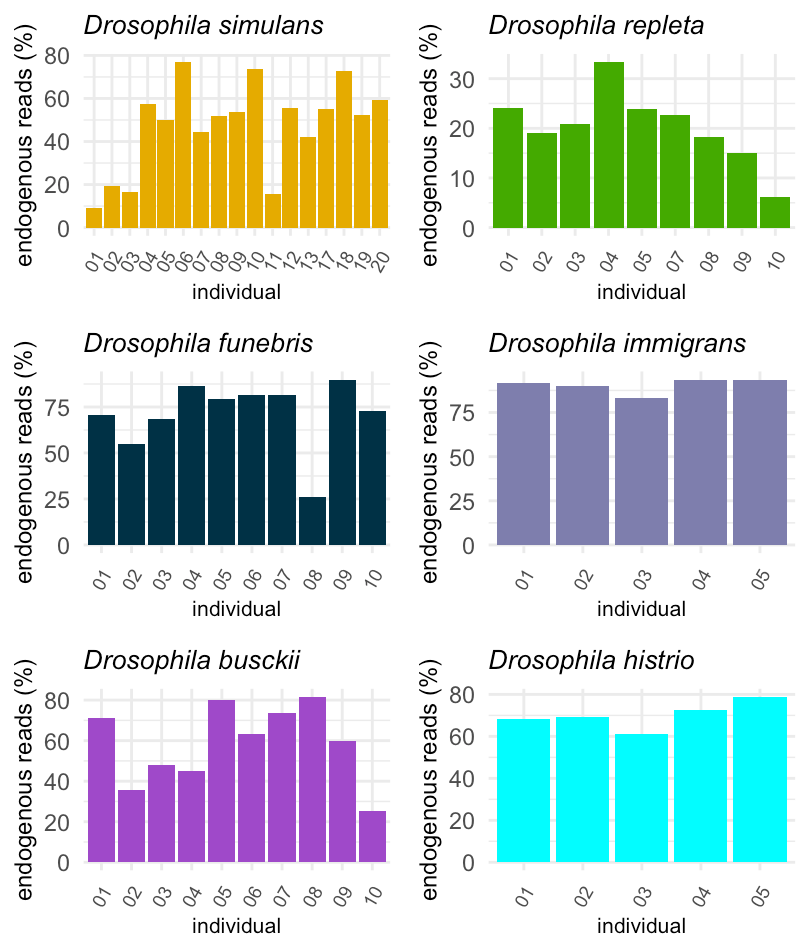
Sequencing: retrieved target genome data
Following the fraction of endogenous reads from multiple specimens of D. simulans (Smithonian National Museum of Natural History – Washington DC), D. repleta (NHM Vienna), D. funebris (NHM Vienna), D. immigrans (NHM Vienna), D. busckii (NHM Vienna) and D. histrio (NHM Vienna). The specimens were collected between 1850 and 1970. The figure demonstrates that our protocol for DNA extraction works perfectly. We obtain sufficient amounts of DNA for sequencing the entire genomes of historical specimens.